Spanish flu tissue of diseased yields new clues about the 1918 epidemic
by
Kai Kupferschmidt and commentaries by Robert Gorter, MD, PhD.
May. 17th, 2021
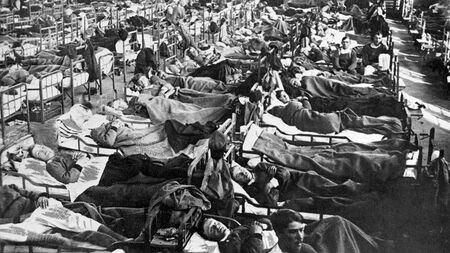
A gymnasium in Boden, Sweden, is filled with pandemic influenza patients in 1918. Researchers have pieced together a full genetic sequence of the flu virus circulating in Europe at the time. (EVERETT COLLECTION/NEWSCOM)
See also:
http://robert-gorter.info/spanish-flu-pandemic/
http://robert-gorter.info/mask-covers-mouth-nose-free-undesirable-side-effects/
On 27 June 1918, two young German soldiers—one age 18, the other 17—died in Berlin from a new influenza strain that had emerged earlier that year. Their lungs ended up in the collection of the Berlin Museum of Medical History, where they rested, fixed in formalin, for 100 years. Now, researchers have managed to sequence large parts of the virus that infected the two men, giving a glimpse into the early days of the most devastating pandemic of the 20th century. The partial genomes hold some tantalizing clues that the infamous flu strain may have adapted to humans between the pandemic’s first and second waves.
The researchers also managed to sequence an entire genome of the pathogen from a young woman who died in Munich at an unknown time in 1918. It is only the third full genome of the virus that caused that pandemic and the first from outside North America, the authors write in a preprint posted on bioRxiv.
“It’s absolutely fantastic to work,” says Hendrik Poinar, who runs an ancient DNA lab at McMaster University. “The researchers have made reviving RNA viruses from archival material an achievable goal. Not long ago this was, like much ancient DNA work, a fantasy.”
Sequencing viral genomes have become routine. In the ongoing coronavirus pandemic, researchers have amassed a database of more than 1 million genomes of SARS-CoV-2, allowing them to watch variants appear and spread while old ones disappear. But few sequences exist of the H1N1 influenza virus that caused the pandemic of 1918–19. In the early 2000s, scientists in the United States painstakingly pieced together one genome from samples taken from a woman’s body buried and preserved in the frozen ground in Alaska. And in 2013 they presented a second genome from a U.S. flu fatality, teased out from autopsy tissue that had been preserved in formalin at the Armed Forces Institute of Pathology. Both studies were time-consuming, costly efforts that few people tried to emulate, says virologist Angela Rasmussen of the Vaccine and Infectious Disease Research Organization at the University of Saskatchewan. Tracking down archived tissue samples is itself a challenge, says evolutionary biologist Michael Worobey of the University of Arizona, a co-author on the new preprint. “It’s all about finding samples,” Worobey says. “Our group has scoured a lot of different locations, and they’re hard to come by.”
Evolutionary biologist Sébastien Calvignac-Spencer of the Robert Koch Institute and his colleagues have now investigated 13 lung tissue samples from between 1900 and 1931 that were in the medical museum in Berlin and in a collection in Vienna. They found bits of RNA from the flu virus in three of them, all dating to 1918. (Like SARS-CoV-2, the influenza virus genome is composed of RNA, not DNA.) Although the RNA was broken down into tiny fragments, there were enough of these to reconstruct the entire genome of the virus from the woman, who was just 17 years old, and close to 90% and 60%, respectively, of the virus that killed the two soldiers. Sequencing genetic material from formalin-fixed tissue is still harder than with other kinds of specimens, Calvignac-Spencer says. “But it’s not the kind of impossible work that we once thought it was.”
The partial genomes from the two soldiers are from the first, milder wave of the pandemic, which was followed by a more severe one that swept the world in the fall of 1918. Scientists have speculated that the virus originated in birds and became better adapted to humans between the first and second waves. One way this could happen is if the gene for haemagglutinin; an important protein on the surface of the virus, underwent an amino acid–swapping mutation that replaced a particular glycine, more often seen in bird flu viruses, with an aspartic acid, which is more characteristic of human viruses. Both German sequences carried an aspartic acid at the position, however, making that scenario unlikely.
The researchers did find an evolutionary clue in the gene for the virus’ nucleoprotein, a structural protein that helps determine what species the virus can infect. The previously reported 1918 flu strains, both from late in the pandemic, carry two mutations in this gene that help influenza avoid the human body’s innate antiviral defenses; the German soldiers’ sequences were more birdlike. “It could be a sign that the virus was evolving to better avoid the human immune response in the first months of the pandemic,” Calvignac-Spencer says. The Munich woman’s flu strain also carried the more birdlike version of the nucleoprotein but given her uncertain date of death, nothing can be concluded about the strain’s evolution.
The full genome from the women yielded other clues, however. The researchers used its genes to resurrect the virus’s polymerase complex, machinery consisting of three proteins that together copy the pathogen’s genome. In cell culture experiments, they discovered the complex from the Munich strain was about half as active as the polymerase complex from the Alaska strain. (The study did not pose safety concerns because the team didn’t reconstitute the whole virus.)
Extrapolating from petri dish studies to human infections is difficult, Poinar says. Still, “The fact that you can test, in vitro, the effects of an ‘extinct’ strain has huge implications in understanding the evolution of virulence and possible countermeasures should we encounter another flu epidemic.”
The work also shows that pathology archives are “treasure troves” that can still yield more information about the 1918 pandemic, Rasmussen says: “If the last 18 months have demonstrated anything, it’s that we would do well to remember the lessons of past pandemics as we try to prevent future ones.”
Posted in: Health doi:10.1126/science.abj5064

Kai Kupferschmidt
Kai Kupferschmidt is a contributing correspondent for Science magazine based in Berlin, Germany. He writes about infectious diseases as well as food science, nutrition, evolution, and science policy. Kai received a diploma in molecular biomedicine from the University of Bonn, Germany, and later visited the Berlin Journalism School.
In 2013 Kai won the Journalism Prize of the German AIDS Foundation. He is the author of a book about the color blue, published in 2019.